Description
RiboCop rRNA Depletion Kits
| This product can be used for SARS-CoV-2 research. |
RiboCop rRNA Depletion Kits for Human/Mouse/Rat and for Bacteria enable removal of ribosomal RNA from total RNA and are suited for Next Generation Sequencing as well as other demanding RNA analysis applications.
New! RiboCop HMR V2 with improved performance on degraded RNA samples.
New! Now also available for combined rRNA and globin mRNA depletion.
Broad Species and Sample Compatibility
RiboCop rRNA Depletion Kits are available for Human/Mouse/Rat (Cat. No. 144), Human/Mouse/Rat plus Globin (Cat. No. 145), and for Bacteria (Cat. No. 125 – 127). For RiboCop HMR compatibility with other species, please consult the FAQs, or contact us. For RiboCop for Bacteria, a dedicated Selection Tool is available.
Efficient Elimination of rRNA
RiboCop allows efficient removal of rRNAs. RiboCop for Human/Mouse/Rat (HMR, Cat. No. 144) removes cytoplasmic 28S, 18S, 5.8S, 45S, 5S, as well as mitochondrial mt16S, mt12S ribosomal RNA sequences in human, mouse, and rat samples. RiboCop for Human/Mouse/Rat plus Globin (HMR+Globin, Cat. No. 145) depletes both rRNA and globin mRNA from blood samples. RiboCop for Bacteria (META, G-, G+, Cat. No. 125 – 127) removes 23S, 16S, and 5S rRNA from Gram positive and Gram negative bacterial species as well as from mixed bacterial samples. Sequencing depth is thus preserved for relevant RNA types.
Simple Workflow
The RiboCop protocol takes 1.5 hours to complete (30 minutes hands-on time). No enzymatic reactions or mechanical shearing steps are involved, leaving full-length transcripts intact for downstream processing.
Flexible Input Requirements
RiboCop is suitable for rRNA depletion of 1 – 1,000 ng total RNA input amounts and is compatible with intact or degraded RNA (including FFPE) samples. Specific input recommendations apply for certain sample types and are described in the respective User Guides.
Automation
Magnetic beads are used for depletion and purification steps rendering the protocol completely automation-friendly.
Various Downstream Applications
RiboCop rRNA Depletion Kits are offered as stand-alone kits in different sizes for use with any downstream application. For convenience, RiboCop for Human/Mouse/Rat Kits (HMR and HMR+Globin) are also available in combination with the CORALL Total RNA-Seq Library Prep Kit (Cat. No. 146 and 147).
Workflow
Samples are treated using a set of affinity probes for specific depletion of rRNA sequences. RiboCop probes efficiently remove rRNA and therefore afford a comprehensive view of transcriptome composition.
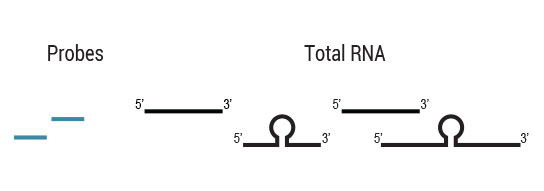
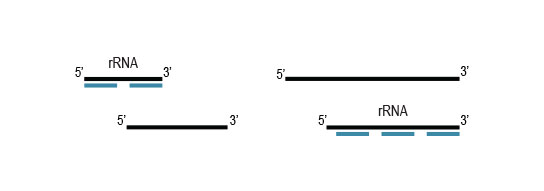
to remove probes along with hybridized
ribosomal RNA from solution.
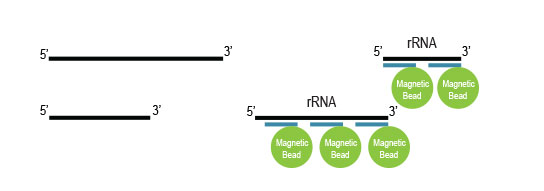
The depleted RNA is purified for downstream
processing.
RiboCop for HMRV2, HMR + Globin & Bacteria
RiboCop rRNA Depletion Kit HMR V2

- Depletion of all cytoplasmic and mitochondrial rRNA from human,
mouse, or rat samples - NEW! Innovative probe design minimizing off-target effects
- 1 ng to 1 µg total RNA input (FFPE compatible)
RiboCop for Human/Mouse/Rat V2
Total RNA from mammalian species is comprised of large amounts of undesired ribosomal RNA (rRNA) which accounts for ~80 – 90 % of all RNA. Lexogen’s RiboCop rRNA Depletion Kits for Human/Mouse/Rat remove undesired cytoplasmic (28S, 18S, 5.8S, and 45S rRNA) and mitochondrial rRNA (mt16S, mt12S, and 5S) from intact as well as degraded material, including formalin-fixed, paraffin-embedded (FFPE) samples.
RiboCop for Human/Mouse/Rat (HMR) V2 is offered as a stand-alone (Cat. No. 144) or as a bundled version with the CORALL Total RNA-Seq Library Prep Kit (Cat. No. 146). RiboCop Kits are compatible with all random primed total RNA library prep kits.
Robust Performance Over a Wide Range of Input Amounts
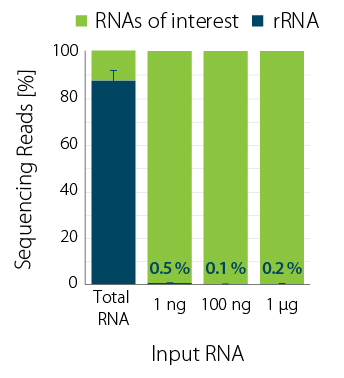
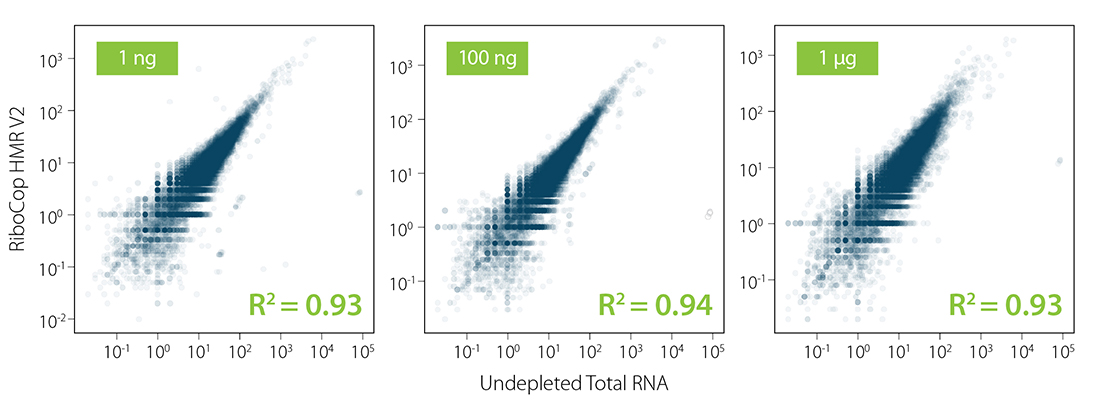
Ribosomal RNA was depleted from Universal Human Reference RNA (UHRR) using RiboCop for Human/Mouse/Rat (HMR) V2 over a wide range of input amounts (1 ng to 1 µg). RiboCop HMR V2 efficiently reduces rRNA reads to below 1% (Fig. 1) while maintaining unbiased transcript abundance (Fig. 2).
Figure 1 | RiboCop rRNA Depletion for Human/Mouse/Rat efficiently removes rRNA across a wide range of input amounts. NGS libraries were prepared using Lexogen’s CORALL Total RNA-Seq Library Prep Kit. Successful depletion was monitored by sequencing (NextSeq500, 1×75 bp) and subsequent analysis of remaining rRNA reads from untreated (Total RNA) and depleted UHRR (1, 100 ng, and 1 µg). The percentage of reads mapping to rRNA is plotted in blue.
Figure 2 | RiboCop maintains unbiased expression profiles while efficiently removing undesired ribosomal RNA from UHRR. Correlation of transcript abundance in untreated samples vs. samples depleted with RiboCop for Human/Mouse/Rat (HMR) V2 for 1 ng – 1 µg UHRR. Libraries were prepared and sequenced as described in Figure 1. Reads were mapped against the GRCh38.95 reference genome using STAR aligner and counted with FeatureCounts (including multi mappers).
RiboCop Performance Across Species
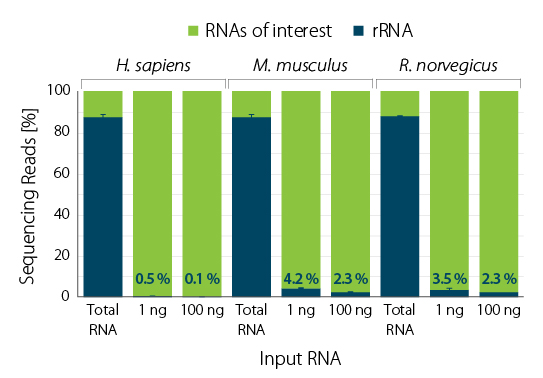
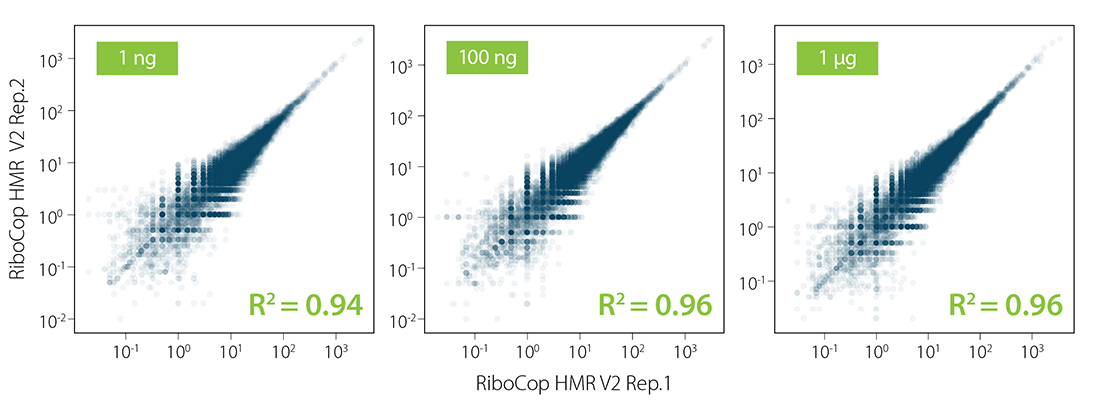
RiboCop enriches RNAs of interest across a wide range of total RNA input amounts from human, mouse, and rat samples (Fig. 3), and shows excellent reproducibility between replicates (Fig. 4).
Figure 3 | RiboCop rRNA Depletion for Human/Mouse/Rat (HMR) V2 removes rRNA from human (UHR), mouse (liver), and rat (liver). Two RNA input amounts of the indicated species were depleted with RiboCop HMR V2. Library preparation and sequencing were performed as described in Figure 1. Reads were mapped against the respective reference genomes for human (GRCh38.95), mouse (mmu_GRCm38.95), and rat (rno_Rnor_6.0.95). The percentage of reads mapping to rRNA is plotted in blue.
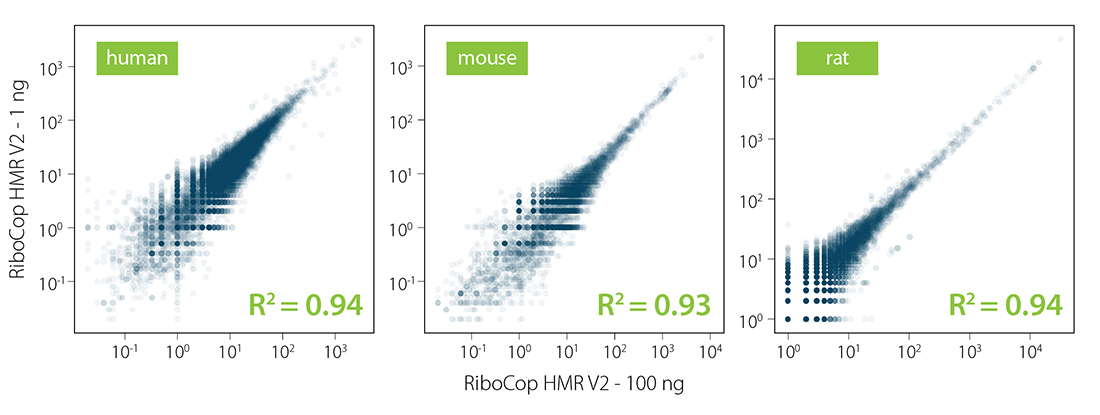
Figure 4 | Excellent reproducibility between replicates. Correlation of replicates for two independent depletion reactions using 1 ng, 100 ng, and 1 µg total UHRR as input. Total RNA was ribo-depleted with RiboCop HMR V2. Library preparation, sequencing, and data analysis were performed as described in Figure 2. Reads were mapped against the GRCh38.95 reference genome.
Excellent Reproducibility and Depletion Without Off-target Effects
RiboCop protocols are robust and highly reproducible. Correlation plots show consistent results over a broad range of inputs (Fig. 5). Additionally, Figure 2 shows a high correlation between transcript abundance in untreated vs. ribo-depleted samples for a wide input range (1 ng to 1 µg) indicating that depletion with RiboCop HMR V2 does not affect the abundance of non-targeted transcripts.
Figure 5 | RiboCop HMR V2 maintains constant transcript abundance across various input amounts. Correlation of transcript abundance in samples depleted with RiboCop HMR V2 for 100 ng RNA vs. 1 ng RNA input for human (UHRR), mouse (mouse liver RNA), and rat (rat liver RNA) samples. Library preparation, sequencing, and data analysis were performed as described in Figure 2. Reads were mapped against the respective reference genomes for human (GRCh38.95), mouse (mmu_GRCm38.95), and rat (rno_Rnor_6.0.95), respectively.
Catalog Nr. |
Product Name |
144.24* |
RiboCop rRNA Depletion Kit for Human/Mouse/Rat (HMR) V2, 24 preps |
144.96* |
RiboCop rRNA Depletion Kit for Human/Mouse/Rat (HMR) V2, 96 preps |
RiboCop rRNA Depletion Kit HMR plus Globin

- Depletion of all cytoplasmic and mitochondrial rRNA
- NEW! Combined rRNA and globin mRNA depletion
- NEW! Innovative probe design minimizing off-target effects
- 1 ng to 1 µg total RNA input (FFPE compatible)
RiboCop for Human/Mouse/Rat plus Globin
Total RNA from mammalian blood is comprised of highly abundant undesired RNA species, such as ribosomal RNA (rRNA), accounting for ~80 – 90 % of total RNA, and globin mRNA, representing 30 -80 % of all mRNAs. Lexogen’s RiboCop rRNA Depletion Kit for Human/Mouse/Rat plus Globin removes undesired cytoplasmic (28S, 18S, 5.8S, and 45S rRNA) and mitochondrial rRNA (mt16S, mt12S, and 5S), and globin mRNA from intact as well as degraded whole blood RNA.
RiboCop for Human/Mouse/Rat plus Globin (HMR+Globin) is offered as a stand-alone kit (Cat. No. 145) or as a bundled version with the CORALL Total RNA-Seq Library Prep Kit (Cat. No. 147). RiboCop Kits are compatible with all random primed total RNA-Seq library prep kits.
Globin Depletion Increases Gene Detection
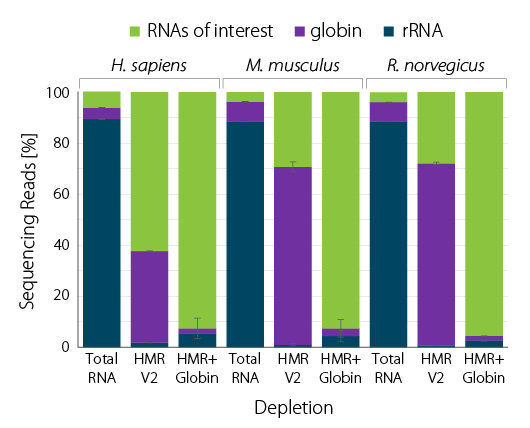
RiboCop for Human/Mouse/Rat plus Globin (HMR+Globin) simultaneously depletes rRNA and globin mRNA, providing a highly convenient workflow for blood samples and freeing up sequencing space for RNAs of interest (Fig. 1). Thus, removal of globin mRNA leads to a significant increase in gene detection (Fig. 2).
Figure 1 | RiboCop rRNA Depletion for Human/Mouse/ Rat plus Globin (HMR+Globin) efficiently removes rRNA and globin mRNA from human blood RNA. A) RNA was extracted from fresh human donor blood using SPLIT RNA Extraction. 10 ng whole blood RNA were depleted with RiboCop HMR V2 or HMR+Globin. Libraries were prepared with the CORALL Total RNA-Seq Library Prep Kit and sequenced on NextSeq500 (1×75 bp). Reads were mapped against the human reference genome (GRCh38.95) and reads mapping to rRNA (blue) and globin (purple) were calculated. B) Correlation plot for depleted blood samples shown in A (uniquely mapping reads). Major globins (marked in red) are retained in samples depleted with HMR V2 and efficiently removed by RiboCop HMR+Globin depletion.
Figure 2 | Increased gene detection upon globin depletion. A) The number of detected genes per number of reads uniquely mapping to exons (FeatureCounts) was plotted for the samples shown in Figure 1. B) Bar plot illustrating the number of detected genes for the samples shown in Figure 1 at 1 M reads /sample, normalized to Counts Per Million (CPM) at a threshold of CPM > 1.
RiboCop Performance Across Species
RiboCop enriches RNAs of interest by efficiently removing rRNA and globin mRNA from human, mouse, and rat whole blood RNA (Fig. 3).
Figure 3 | RiboCop rRNA Depletion for Human/Mouse/ Rat plus Globin (HMR+Globin) enriches for RNAs of interest in blood sample across species. 100 ng whole blood RNA were depleted with either RiboCop HMR V2 or RiboCop HMR+Globin. Library preparation, sequencing, and data analysis was performed as described in Figure 1. Reads were mapped against the respective reference genomes for human (GRCh38.95), mouse (mmu_GRCm38.95), and rat (rno_Rnor_6.0.95), respectively.
Catalog Nr. |
Product Name |
145.24 |
RiboCop rRNA Depletion Kit for Human/Mouse/Rat plus Globin (HMR+Globin), 24 preps |
145.96 |
RiboCop rRNA Depletion Kit for Human/Mouse/Rat plus Globin (HMR+Globin), 96 preps |
RiboCop rRNA Depletion Kits for Bacteria

- Depletion of 23S, 16S, and 5S rRNA
- Optimized Probe Mixes for monocultures or mixed communities
- Innovative probe design minimizing off-target effects
- 1 ng to 1 µg total RNA input (FFPE compatible)
RiboCop for Bacteria
RNA extracted from bacterial species comprises up to 98 % of the ribosomal RNAs (rRNA), presenting a unique challenge especially when analyzing the transcriptome capacity from complex bacterial communities. Lexogen’s RiboCop rRNA Depletion Kits for Bacteria efficiently remove 23S, 16S, and 5S rRNA from mixed bacterial samples and monocultures.
RiboCop is offered as a stand-alone kit with the option to choose from three optimized Probe Mixes for depletion of rRNA from mixed bacterial samples (META, Cat. No. 125) or from Gram negative or Gram positive bacteria (G- or G+, Cat. No. 126 and 127, respectively) grown in monoculture.
RiboCop is compatible with all random primed library prep kits such as the CORALL Total RNA-Seq Library Prep Kit (Cat. No. 095)or adapter ligation protocols such as the Small RNA-Seq Library Prep Kit (Cat. No. 052).
Robust Performance Over a Wide Range of Input Amounts while Maintaining Unbiased Transcript Abundance
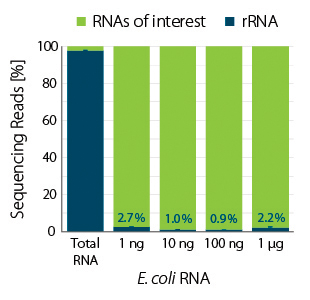
Ribosomal RNA was depleted from E. coli total RNA using RiboCop for Gram Negative Bacteria over a wide range of input amounts (1 ng to 1 µg). RiboCop for Bacteria efficiently reduces rRNA reads from 98 % to 1-3 % over a broad range of input amounts (Fig. 1) while maintaining unbiased transcript abundance (Fig. 2).
Figure 1 | RiboCop rRNA Depletion for Bacteria efficiently removes rRNA across a wide range of input amounts. NGS libraries were prepared using Lexogen’s CORALL Total RNA-Seq Library Prep Kit. Successful depletion was monitored by sequencing and subsequent analysis of remaining rRNA reads from untreated (Total RNA) and depleted E. coli RNA (1, 10, 100 ng and 1 µg). Sequencing on Illumina NextSeq (1×75 bp) and analysis by mapping to the MG1655 reference using BBMap. The percentage of reads mapping to rRNA is plotted in blue.

Figure 2 | RiboCop maintains unbiased expression profiles while efficiently removing undesired ribosomal RNA. Correlation of transcript abundance in untreated samples vs. samples depleted with RiboCop for Gram Negative Bacteria for 1 ng – 1 µg E. coli RNA input. Libraries were prepared using Lexogen’s CORALL Total RNA-Seq Library Prep Kit and sequenced on Illumina NextSeq (1×75 bp). Reads were mapped against the MG1655 reference genome using STAR aligner and counted with FeatureCounts (including multi mappers).
Performance on Meta-transcriptome Samples
The RiboCop rRNA Depletion Kit for Mixed Bacterial Samples (META) is specifically designed for depletion of complex, mixed populations such as environmental communities or microbiome samples. Low quality microbiome samples from stool extractions were used for characterization of the Probe Mix designed for meta-transcriptomics (Tab. 1). The META Probe Mix can also be used for efficient and robust depletion of monocultures up to 100 ng total RNA input (Fig. 3) while maintaining unbiased transcript expression (Fig. 4).
| Sample | % rRNA reads |
| stool microbiome 10 ng | 14.75 (± 1.4) |
| stool microbiome 100 ng | 15.32 (± 1.4) |
| stool microbiome 1 µg | 23.37 (± 1.6) |
Table 1 | Depletion rates for meta-transcriptome analyses. Different amounts of stool microbiome RNA was treated with RiboCop for Mixed Bacterial Samples (META). NGS-Libraries were prepared, sequenced, and analyzed as described in Fig. 1. Reads were mapped to ~60 annotations.
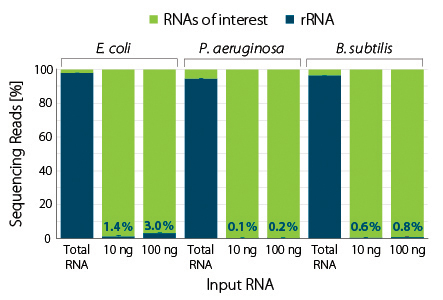
Figure 3 | RiboCop rRNA Depletion for Mixed Bacterial Samples (META) efficiently removes rRNA from various bacterial species. Two RNA amounts from monocultures of the indicated species were subjected to rRNA depletion using the META Probe Mix. Library preparation and sequencing were performed as described in Fig. 1. Reads were mapped against the respective genomes of E. coli MG1655, P. aeruginosa PAO1, and B. subtilis 168. The percentage of reads mapping to rRNA is plotted in blue.

Figure 4 | RiboCop META maintains constant transcript abundance across various species. Correlation of transcript abundance in untreated samples vs. samples depleted with RiboCop for Mixed Bacterial Samples (META) for 100 ng RNA input of three different species. Library preparation, sequencing, and data analysis were performed as described in Fig. 2. Reads were mapped against the E. coli MG1655, P. aeruginosa PA01, or B. subtilis 168 reference genomes, respectively.
We have extensively tested Lexogen’s RiboCop META rRNA depletion kit on bacteria for transcriptomics. Due to a research focus in infection biology at our university, we are working on RNA sequencing projects with various types of bacteria. The RiboCop META is a versatile tool with very good depletion results for all types of bacteria tested so far. Therefore, we would like to continue to use it for future transcriptome profiling projects.
Excellent Reproducibility and Depletion Without Off-target Effects
RiboCop protocols are robust and highly reproducible. Correlation plots show excellent reproducibility between replicates (Fig. 5) independent of the species or the Probe Mix used for depletion.
Figures 2 and 4 additionally show a high correlation between transcript abundance in untreated vs. ribo-depleted samples for a wide input range (1 ng to 1 µg, Fig. 2) as well as across various species (Fig. 4) without outliers. This indicates that depletion with RiboCop for Bacteria occurs without of off-target effects.
Figure 5 | Excellent reproducibility between replicates. Correlation of replicates for two independent depletion reactions using 10 ng total RNA of the indicated species. Total RNA was ribo-depleted with RiboCop for Gram Negative or Gram Positive Bacteria, respectively. Library preparation, sequencing, and data analysis were performed as described in Fig. 4.
RiboCop for Bacteria Selection Tool
If you are uncertain which kit to choose, then the RiboCop for Bacteria Kit Selection Tool can help to find the best-suited kit for your species of interest.
Catalog Nr. |
Product Name |
125.24 |
RiboCop rRNA Depletion Kit for Mixed Bacterial Samples (META), 24 preps |
125.96 |
RiboCop rRNA Depletion Kit for Mixed Bacterial Samples (META), 96 preps |
126.24 |
RiboCop rRNA Depletion Kit for Gram Negative Bacteria (G-), 24 preps |
126.96 |
RiboCop rRNA Depletion Kit for Gram Negative Bacteria (G-), 96 preps |
127.24 |
RiboCop rRNA Depletion Kit for Gram Positive Bacteria (G+), 24 preps |
127.96 |
RiboCop rRNA Depletion Kit for Gram Positive Bacteria (G+), 96 preps |
FAQs
1.1 Do I need to purchase any additional modules or reagents for an efficient rRNA depletion with RiboCop?
80% EtOH is required. All other reagents are provided in the kit.change this text.
1.2 Can I run the protocol without thermomixer?
If you don´t have a thermomixer available you can just run it on a thermocycler without agitation.
1.3 Can I use RiboCop in combination with the CORALL Total RNA-Seq kit?
RiboCop rRNA depleted total RNA is suitable to be used with the CORALL Total RNA-Seq Library Prep Kit. For convenience bundled versions consisting of CORALL Total RNA-Seq Library Prep Kit with RiboCop HMR V2 (Cat. No. 146) or with RiboCop HMR+Globin (Cat. No. 147) are also available.
1.4 Can RiboCop HMR and RiboCop Bacteria be used in combination for a one-step rRNA depletion?
Yes. Please contact info@gomolecular.pk to obtain the protocol for simultaneous host and bacterial rRNA depletion.
2.1 For which species can I use the RiboCop HMR rRNA Depletion Kit?
RiboCop HMR contains probes for the rRNA depletion of human, mouse and rat RNA samples and was also successfully tested on hamster, mini pig, and zebrafish samples.
2.2 What is the expected RNA recovery rate after rRNA depletion?
Depending on input sample, standard recovery rate yield varies from 1 – 3%.
2.3 How much rRNA is left after depletion with RiboCop HMR V2?
RiboCop HMR V2 efficiently reduces rRNA reads from 80 – 90 % down to below 1 % over a broad range of input amounts. Depending on input sample, depletion down to 1 – 2 % is standard.
2.4 What RNA input amounts can be used?
RiboCop HMR is suitable for input amounts from 1 ng – 1 µg of total RNA input.
2.5 What are the differences between RiboCop HMR V1.3 and V2?
The difference between the two versions is the Probe Mix provided with the kit (HMR V2, previously PM in V1.3). In V2, a sophisticated algorithm was utilized to design efficient probes that minimize off-target effects. This new Probe Mix is designed to cover all transcript variants. The workflow and steps follow the streamlined protocol from V1.3. Additionally, enhanced depletion is achieved with V2 compared to V1.3 for both high- and low-quality RNA.
2.6 What sample types was RiboCop HMR+Globin tested on?
RiboCop for Human/Mouse/Rat plus Globin (HMR+Globin) was tested extensively on whole blood RNA of varying quality from human, mouse, and rat.
2.7 What level of depletion does RiboCop HMR+Globin achieve?ed on?
Globin mRNA accounts for 30 – 80 % of blood mRNA. Using RiboCop for Human/Mouse/Rat plus Globin (HMR+Globin), >90 % of the globin mRNA is efficiently removed.
2.8 Why is the percentage of rRNA reads higher when globin is depleted?
This increase is expected due to the fact that 35 – 75 % of mRNA (specifically, globin mRNA) are removed. In absolute numbers, the rRNA stays the same but makes up a larger fraction of reads due to the decrease in non-rRNA fraction.
2.9 How much sequencing depth is saved when depleting globin mRNA?
3.1 How do I select the optimal RiboCop for Bacteria kit?
For rRNA depletion from Gram negative (e.g., E. Coli) or Gram positive (e.g., B. Subtilis) bacterial monocultures we recommend the RiboCop for Gram Negative or Gram Positive Bacteria kits, respectively (Cat. No. 126 or 127).
The META Probe Mix (Cat. No. 125) is primarily suited for depletion of total RNA from mixed bacterial samples (e.g., for meta-transcriptome analysis of complex communities or microbiome samples).
3.2 What RNA input amounts can be used?
The recommended input range for efficient depletion with RiboCop rRNA Depletion Kits for Bacteria is 1 – 1,000 ng of total RNA for monocultures of Gram negative or Gram positive bacteria using the respective G- and G+ Probe Mixes (Cat. No. 126 and 127, respectively). We suggest initial experiments to be performed using 100 ng of input material.
The META Probe Mix (Cat. No. 125) is primarily suited for depletion of 1 – 1,000 ng of total RNA from mixed bacterial samples. The META Probe Mix may also be used at 1 – 100 ng when working with monocultures.
3.3 What level of depletion can be expected from RiboCop for Bacteria?
Processing high quality RNA from bacterial monocultures with the Gram Positive or Gram Negative Probe Mixes usually results in less than 1 % remaining rRNA reads.
Depletion rates for the META Probe Mix using RNA from mixed bacterial samples can vary depending on the sample. For specific questions please contact info@gomolecular.pk
Depletion rates may vary depending on the quality of the RNA and purity of the sample. Contaminants, such as salts, metal ions and organic solvents, may have a negative impact on the efficiency of the protocol.
3.4 Can the META Probe Mix be used with a monoculture samples?
Yes, however the recommended input range in this case is 1 – 100 ng of total RNA.
Downloads
![]() User Guide RiboCop for Human/Mouse/Rat (HMR) V2 – release 21.09.2020
User Guide RiboCop for Human/Mouse/Rat (HMR) V2 – release 21.09.2020
![]() User Guide RiboCop for Bacteria – update 31.08.2020
User Guide RiboCop for Bacteria – update 31.08.2020
![]() Product Flyer
Product Flyer
![]() Technical Note – RiboCop for HMR (Human/Mouse/Rat)
Technical Note – RiboCop for HMR (Human/Mouse/Rat)
![]() Technical Note – RiboCop for Bacteria
Technical Note – RiboCop for Bacteria
RiboCop rRNA Depletion
RiboCop rRNA Depletion Kits (Human/Mouse/Rat)
Use for Cat. No.s: 037 (RiboCop V1.3 Human/Mouse/Rat), 096 (CORALL Total RNA-Seq Kit with RiboCop).
Current Version:
![]() 144UG288V0100 – User Guide RiboCop for Human/Mouse/Rat (HMR) V2 – release 21.09.2020
144UG288V0100 – User Guide RiboCop for Human/Mouse/Rat (HMR) V2 – release 21.09.2020
![]() 144MS076V0500 – MSDS Information for all RiboCop rRNA Depletion Kits – update 21.09.2020
144MS076V0500 – MSDS Information for all RiboCop rRNA Depletion Kits – update 21.09.2020
Previous versions
![]() 037UG073V0300 – User Guide RiboCop for Human/Mouse/Rat (HMR) – update 09.09.2019
037UG073V0300 – User Guide RiboCop for Human/Mouse/Rat (HMR) – update 09.09.2019
![]() 037UG073V0202 – User Guide RiboCop for Human/Mouse/Rat (HMR) – update 29.08.2018
037UG073V0202 – User Guide RiboCop for Human/Mouse/Rat (HMR) – update 29.08.2018
![]() 037UG073V0201 – User Guide RiboCop for Human/Mouse/Rat (HMR) – update 26.07.2017
037UG073V0201 – User Guide RiboCop for Human/Mouse/Rat (HMR) – update 26.07.2017
![]() 037MS076V0400 – MSDS Information for all RiboCop rRNA Depletion Kits – update 27.01.2020
037MS076V0400 – MSDS Information for all RiboCop rRNA Depletion Kits – update 27.01.2020
RiboCop rRNA Depletion Kits for Bacteria
Use for Cat. No.s: 125 (RiboCop rRNA Depletion Kit for Mixed Bacterial Samples (META)), 126 (RiboCop rRNA Depletion Kit for Gram Negative Bacteria (G-)), 127 (RiboCop rRNA Depletion Kit for Gram Positive Bacteria (G+))
Current Version:
![]() 125UG246V0101 – User Guide RiboCop for Bacteria – update 31.08.2020
125UG246V0101 – User Guide RiboCop for Bacteria – update 31.08.2020
Previous versions
![]() 125UG246V0100 – User Guide RiboCop for Bacteria – release 24.02.2020
125UG246V0100 – User Guide RiboCop for Bacteria – release 24.02.2020