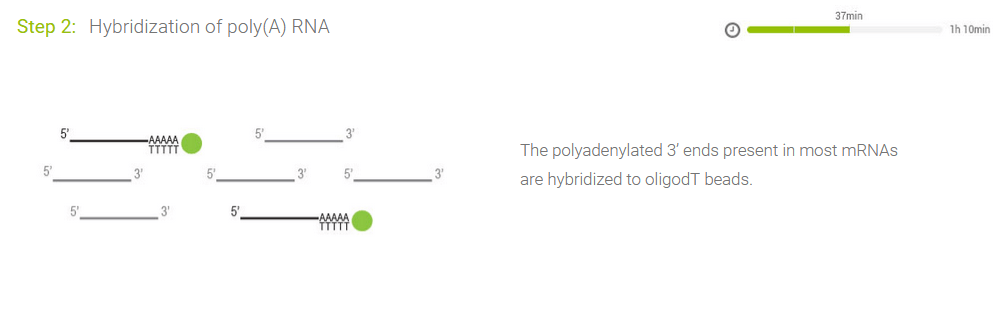Description
The kit is suitable to extract poly(A) RNA from 480 µg of total RNA input (96 reactions with 5 µg total RNA each).
The upgrade V1.5 contains new oligo(dT) Magnetic Beads (MB), compared to the V1.0 kit. The beads and solutions included in the V1.5 kit offer improved performance.
Features and Application:
- RNA library preparation and sequencing (e.g., CORALL mRNA-Seq Library Prep Kit)
- Microarray
- SAGE, RACE
- cDNA synthesis
- cDNA library construction
- RT-PCR and others
Kit Content:
- Buffers and oligo(dT) Magnetic Beads
157.96 (Poly(A) RNA Selection Kit V1.5)
Optimized V1.5 Upgrade
The upgrade V1.5 contains new oligo(dT) Magnetic Beads (MB), compared to the V1.0 kit. The beads and solutions included in the V1.5 kit offer improved performance.
Highly Specific for Poly(A) RNA
Other RNA species (rRNA and tRNA) do not contain poly(A) sequences and therefore will not bind to the oligo(dT) beads.
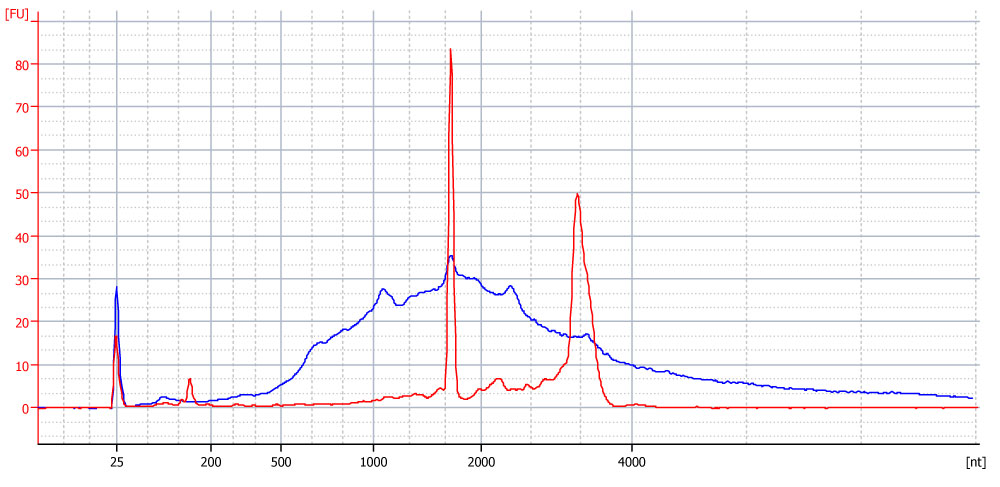
Figure 1. Bioanalyzer traces of Universal Human Reference Total RNA (UHRR, red trace) and poly(A) selected RNA from 5 µg UHRR (blue trace).
Various Downstream Applications
Isolated mRNA can directly be used for RNA-Seq library preparation (e.g., CORALL RNA-Seq Library Prep kits), SAGE, CAGE, cloning, microarrays, cDNA synthesis, and others.
Rapid Turnaround
Polyadenylated RNAs can be isolated from total RNA samples within about one hour.
FAQ
Frequently Asked Questions
Please find a list of the most frequently asked questions below. If you cannot find the answer to your question here or want to know more about our products, please contact info@gomolecular.pk .
1. What is the length of the oligodT?
The oligodT consists of 30 Ts.
2. Which RNAs get isolated using the Poly(A) RNA Selection Module?
The poly(A) RNA Selection Module enriches for all RNAs that have a poly(A) tail, such as mRNA, but also mitochondrial rRNA, which can make up to 4 % of the isolated poly(A) RNA (=0.08% of the total RNA). Therefore, between 2 – 4 % of sequencing reads can correspond to rRNA when using various standard RNA-Seq library preps.
3. What is the difference between V1.0 (039.100) and V1.5 (157.96)?
The Poly(A) RNA Selection Kit V1.5 features new oligo(dT) Magnetic Beads (MB) compared to the V1.0 kit. The V1.5 kit protocol has been carefully optimized to maximize poly(A) RNA recovery. Additional specific changes made in the V1.5 Kit and protocol include:
- Updated Hybridization (HYB) and Bead Wash (BW) buffer compositions (for compatibility with new oligo(dT) Magnetic Beads).
- Reduction of the Magnetic Bead volume to 2 µl (vs. 10 µl in V1.0), which can be used for total RNA inputs up to 5 µg.
4. Do I need two rounds of RNA-binding?
Repeated rounds of poly(A) selection are not required due to the high specificity of the poly(A) selection. One poly(A) selection round is enough. Repeated rounds are often performed to remove unspecifically bound rRNA.
5. What is the poly(A) RNA recovery?
The amount of poly(A) RNA varies depending on the tissue and cell type the total RNA is derived from. Poly(A) RNA usually makes up 1-3 % of the total RNA.
6. After the poly(A) selection the Bioanalyzer still detects some ribosomal peaks? Why is that so?
Agilent’s Bioanalyzer and other electrophoresis platforms often try to detect 18S and 28S rRNA peaks at all costs. Therefore, highly abundant transcripts are sometimes wrongly identified as rRNA.
In the traces below you’ll see that the Bioanalyzer software classifies two peaks within the trace of a poly(A) selected sample as 18S and 28S rRNA (Figure A). However, the overlay with diluted total RNA clearly demonstrates that those peaks are not rRNA peaks (Figure B). Poly(A) selection was successful and no rRNA was left. This was also confirmed by NGS sequencing.
Figure A. Bioanalyzer trace after the Poly(A) RNA Selection using 5 µg of Universal Human Reference RNA (UHRR). The Bioanalyzer software wrongly classifies two peaks within the trace of the poly(A) selected sample as 18S and 28S rRNA.
Figure B. Overlay of Bioanalyzer traces before and after the Poly(A) RNA Selection. The overlay with diluted total RNA clearly demonstrates that those peaks identified as rRNA (in A) are not rRNA peaks.
Red trace:
diluted total Universal Human Reference RNA (UHRR), intact (RIN 8.5).
Blue trace:
poly(A) RNA selected RNA.
7. How can I scale up the Poly(A) Selection reaction for input amounts higher than 5 µg?
Increasing the proportional bead volume to suit the input RNA amount will enable poly(A) selection from higher inputs. For recommendations based on your intended input please contact info@gomolecular.pk .
Downloads
Use for Cat. No.: 157 (Poly(A) RNA Selection Kit V1.5).
![]() 157UG069V0100 – User Guide – update 11.11.2020
157UG069V0100 – User Guide – update 11.11.2020
![]() 157MS078V0100 – MSDS Information for Poly(A) RNA Selection Kit – release 08.10.2020
157MS078V0100 – MSDS Information for Poly(A) RNA Selection Kit – release 08.10.2020