Description
| Illumina Instruments | Workflow | Cat. No. |
| NovaSeq™ 6000 (v1.0 reagent kits)HiSeq® 3000/4000 (SR)HiSeq® 2000/2500MiSeq® | A | 107-110.96 and 120.384113.96, 129-131.96, and 115.384 (QuantSeq FWD)117.96, 132-134.96, and 119.384 (CORALL Total)158-161.96 and 163.384 (CORALL mRNA) |
| NovaSeq™ 6000 (v1.5 reagent kits)NextSeq™ 500/550HiSeq® 3000/4000 (PE)MiniSeq™ 6000iSeq™ 100 | B | 111.96114.96 (QuantSeq FWD)118.96 (CORALL Total)162.96 (CORALL mRNA) |
With the UDIs Add-on kits UDIs are introduced at the PCR step of following library preparation kits: QuantSeq 3’ mRNA-Seq FWD and REV (Cat. No. 015 and 016), QuantSeq-Flex (Cat. No. 033, 034, 035), CORALL Total RNA-Seq (Cat. No. 095), and others.
The UDI 12 nt Unique Dual Indexing Sets are also available in the QuantSeq-FWD (Cat. No. 113 – 115 and 129 – 131), CORALL Total (Cat. No. 117 – 119 and 132 – 134), and CORALL mRNA-Seq (Cat. No. 158 – 163) with UDI kits.
- Kit features:
- Up to 384 Unique Dual Indices (UDIs) for high-level multiplexing
- 12 nt long indices with read-out options of 12, 10, and 8 nt
- Optimized sets for different workflows on Illumina machines
- Maximal inter-index distance for each sample size enables efficient error correction
- Demultiplexing and Error Correction Tool – iDemux
- Maximizing sequencing data output
- Kit size: 96 or 384 rxns of each UDI (1 rxn/UDI)
- Compatibility information
- Product: Lexogen’s CORALL Total RNA-Seq, CORALL mRNA-Seq, and QuantSeq, as well as any RNA and DNA sequencing library prep methods utilizing TruSeq™ – compatible stubby adapters (where partial Illumina adapters are introduced during the workflow and completed with the index information during the library amplification step).
- Platform: Illumina
Lexogen UDI 12 nt Unique Dual Indexing Add-on Kits
107 (Lexogen UDI 12 nt Add-on Kit, Set A1),
108 (Lexogen UDI 12 nt Add-on Kit, Set A2),
109 ((Lexogen UDI 12 nt Add-on Kit, Set A3),
110 (Lexogen UDI 12 nt Add-on Kit, Set A4),
111 (Lexogen UDI 12 nt Add-on Kit, Set B1).
NOTE: If you have purchased a CORALL or QuantSeq Kit with UDI 12 nt, this Instruction Manual is not required in addition (all protocol information is contained in the CORALL or QuantSeq with UDI full User Guides).
Lexogen i5 6 nt Dual Indexing Add-on Kits
No.s: 047.4×96 (Lexogen i5 6 nt Dual Indexing Add-on Kit),
047.96 (Lexogen i5 6 nt Unique Dual Indexing Add-on Kit)
Lexogen i1 12 nt RT-Set for QuantSeq-Pool
Use for Cat. No: 139.96 (QuantSeq-Pool Sample-Barcoded 3′ mRNA-Seq Library Prep Kit for Illumina).
Lexogen 12 nt Unique Dual Index System (UDI) for RNA-Seq
Lexogen’s UDI 12 nt Unique Dual Indexing Sets feature superior error correction for maximal sequencing data output and are introduced at the PCR step of Lexogen’s library kits. Convenient bundles containing the 12 nt UDIs are available for:
- QuantSeq 3’ mRNA-Seq Library Prep Kit FWD for Illumina
- CORALL Total RNA-Seq Library Prep Kit for Illumina
- CORALL mRNA-Seq Library Prep Kit for Illumina
Lexogen UDI 12 nt Unique Dual Indexing Add-on Kits (Cat. No. 107-111.96 and 120.384) are available to replace the provided single indexing system (i7 6 nt) in the following Lexogen library prep kits:
- QuantSeq 3’ mRNA-Seq Library Prep Kit REV for Illumina
- QuantSeq-Flex Targeted RNA-Seq Library Prep Kit V2 for Illumina
The 12 nt UDI Add-on kits are also compatible with other vendor’s RNA-Seq library prep protocols.
The UDI 12 nt Unique Dual Indexing Sets (Cat. No. 101-105.96 and 156.384) containing only primer plates without PCR enzymes are available only for use with:
- QuantSeq-Pool Sample-Barcoded 3’ mRNA-Seq Library Prep Kit for Illumina
- LUTHOR 3’ mRNA-Seq Library Prep for Illumina
Introduction
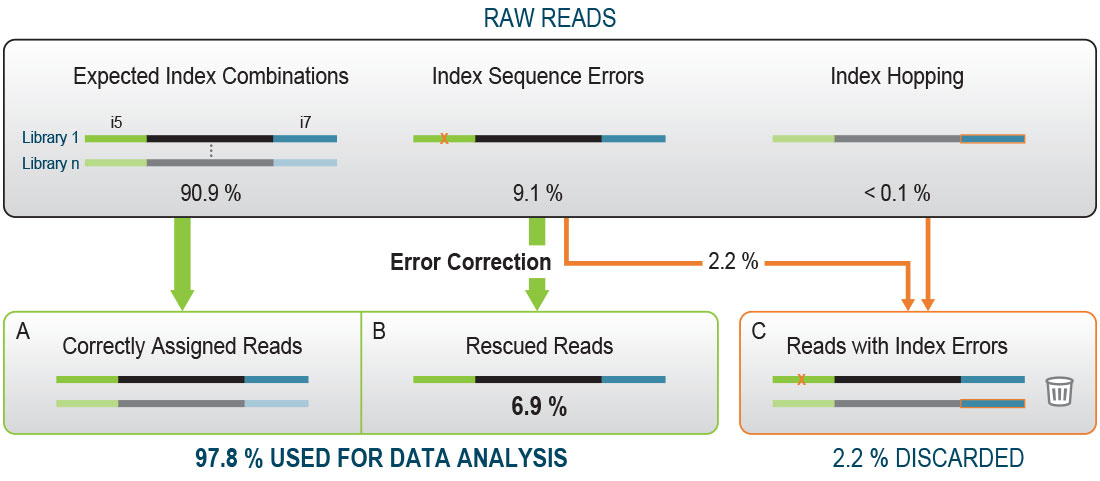
A critical consideration for any multiplexed RNA-Seq workflow is to avoid errors in the index read-out, which can result in the mis-assignment of sequencing reads to the wrong samples. While the majority of the raw reads will have the expected index combinations (Fig. 1A), read mis-assignment can occur on all Illumina platforms. This happens due to two main events: Index Hopping and random Index Sequence Errors.
During Index Hopping an index sequence of one library is incorrectly added to another library which may affect 0.1 – 2 % of all reads [1]. Only the use of Unique Dual Indexing (UDI), where each library in a given pool is barcoded with unique i7 and unique i5 index sequences, unambiguously identifies reads with hopped indices. Such reads are removed from downstream analysis and discarded (Fig. 1C).
Read mis-assignment due to random Index Sequence Errors occurs when an error in one index sequence transforms the index into another one that is present within the same multiplexed sample pool. UDIs resolve such mis-assignment and the read is discarded.
More frequently, an Index Sequence Error results in an index sequence that does not match any other index in the pool, and the read is initially classified as undetermined. If the index sequence in question is different enough from the other index sequences in this pool, then error correction can be applied to recover a significant share of these reads (4 – 7 % of the initial reads, Fig. 1B). The performance of this error correction depends predominantly on the quality of the index design, as deficient index design can result in a higher rate of faulty error correction. Due to their unique design the Lexogen UDI 12 nt Unique Dual Indices minimize the impact of Index Sequence Errors and enable maximal data output gain by error correction.
References
[1] Illumina, Effects of Index Misassignment on Multiplexing and Downstream Analysis (2017) 770-2017-004-D.
Figure 1 | The effects of Index Hopping and Index Sequence Errors in a pool of libraries with Unique Dual Indexing. Read mis-assignment caused by Index Hopping can be avoided by using Unique Dual Indexing (UDI). Reads with hopped indices are irreversibly discarded (C). Reads with random Index Sequence Errors resulting in an index not present in the pool are classified undetermined. Accurate error correction can rescue most of these reads making them available for downstream data analysis (B). The percentage values were derived from an RNA-Seq experiment pooling 96 libraries with Lexogen’s 12 nt UDIs and full 12 nucleotide index read-out on an Illumina NextSeq500.
Universal Application
The Lexogen UDI 12 nt Unique Dual Indexing Add-on Kits are compatible with library prep kits for RNA and DNA sequencing from all vendors utilizing TruSeq™ – compatible stubby adapters (where partial Illumina adapters are introduced during the workflow and completed with the index information during the library amplification step).
Superior Error Correction Maximizes Sequencing Yield
The Lexogen UDI 12 nt Unique Dual Indices are 12 nucleotides (nt) long and designed to maximize inter-index distance for different sample numbers and index read-out lengths. In a typical experiment using the full 12 nt index read-out around 9.1 % of the initial raw reads contain a random Index Sequence Error (Fig. 2A). This renders them undetermined, hence removing these reads from downstream analysis.
Lexogen’s advanced index design enables the rescue of 76 % of these undetermined reads (6.9% of the initial reads), even if multiple nucleotides of the index contain errors. The useful output thereby increases to 97.8 % of the initial reads, an unprecedented performance due to the cutting-edge index design (Fig. 2B).
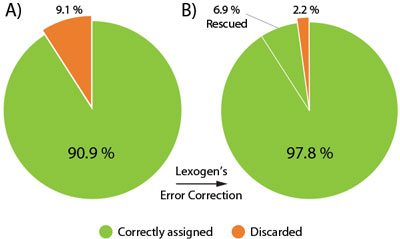
Figure 2 | Maximizing read output with Lexogen’s 12 nt UDIs and error correction. 96 multiplexed libraries were sequenced on an Illumina NextSeq500 with 12 nt UDI read-out. A) In a standard RNA-Seq experiment a significant number of reads is undetermined (orange) due to random Index Sequence Errors. B) Lexogen’s 12 nt Unique Dual Indices are optimized for maximal error correction with highest accuracy. Lexogen’s Error Correction Tool allows almost 7% of originally undetermined reads to be confidently rescued and correctly assigned to the respective library.
Scalable Index Read-out Length
The design of Lexogen’s 12 nt UDIs enables scalable read-out lengths of 12, 10, and 8 nucleotides. The UDIs therefore support all kinds of requirements for multiplexing, which depend on experiment type, sequencing equipment, desired read depth, and / or the number of pooled libraries. For small sample sizes (e.g., 24 samples) short indices (e.g. 8 or 10 nt) are sufficient to ensure high accuracy and reliable error correction. For more than 96 samples however, 8 nt index read-out does not allow reliable error correction anymore, and 10 or 12 nt read-outs are required.
While needing slightly more sequencing cycles, 12 nt long index sequences also provide the ability to correct not only one but two (or in very small sets, three) Index Sequence Errors. Adjustable index read-out-length allows tuning your indexing needs to the experiment design, without the need to purchase separate indexing sets.
Nested Index Set Design for Highest Accuracy
To provide optimal index subsets for these varying multiplexing needs, Lexogen has designed the 12 nt UDIs in a nested approach: Small subsets benefit from Lexogen’s nested index system by having the largest inter-index distance and highest error correction capacity while larger subsets provide for higher multiplexing needs.
Moreover, all subsets are nucleotide-balanced at each index position for optimized cluster identification in the NGS run. Using a proprietary algorithm, Lexogen has designed more than 9,216 UDIs (24x 384 subsets) with the capacity of correcting at least one error. Such sets with more than 384 UDIs are available upon request and enable extreme levels of multiplexing while still providing excellent error correction.

Figure 3 | Distance and error correction in Lexogen’s nested 12 nt UDI sets. Illustration of inter-index distance (D) and number of possible error corrections (ec) in a nested index set with 12 nt read-out. An optimized set of 384 indices contains a subset of 96 indices with larger distances and enhanced error correction. Within these 96 indices, a subset of 24 indices is optimized even further, while a 4 index subset features the highest possible inter-index distance and error correction capacity.
Conclusion
The Lexogen UDI 12 nt Unique Dual Index system adapts to the user’s needs while always providing highest inter-index distance and maximal error correction capacity. Read mis-assignment due to Index Hopping is avoided, and Index Sequence Errors can be corrected with highest accuracy. Thereby, the system provides the optimized indexing solution for current and future barcoding requirements.
| Subset | 12 nt | 10 nt | 8 nt | |||
| D | ec | D | ec | D | ec | |
| 384 | 4 | 1 | 3 | 1 | 2 | 1* |
| └ 96 | 5 | 2 | 4 | 1 | 3 | 1 |
| └ 24 | 6 | 2 | 5 | 2 | 4 | 1 |
| └ 4 | 7 | 3 | 6 | 2 | 5 | 2 |
Table 1 | Comparison of distance and error correction capacity in Lexogen’s nested 12 nt UDI sets with 8, 10, and 12 nt read-out. Inter-index distance (D) and number of errors that can be corrected (ec) are compared for subsets of 384, 96, 24, and 4 libraries and the three possible read-out lengths. For smaller subsets (up to 96 samples) a read-out of 8 or 10 nt allows correction of one error and thus recovery of additional reads. Larger subsets require a read-out of 10 or 12 nt to benefit from the error correction. The ec values represent the number of all errors (including substitutions, insertions, and deletions) that can be confidently corrected, except for *. In this case error correction can only address substitutions.
Downloads
Lexogen 12 nt Unique Dual Index System (UDI) for RNA-Seq
![]() User Guide for Lexogen 12 nt Unique Dual Indexing Add-on Kit – update 23.02.2021
User Guide for Lexogen 12 nt Unique Dual Indexing Add-on Kit – update 23.02.2021
![]() User Guide for QuantSeq 3‘ mRNA-Seq Library Prep Kit FWD with Unique Dual Indices – release 27.12.2019
User Guide for QuantSeq 3‘ mRNA-Seq Library Prep Kit FWD with Unique Dual Indices – release 27.12.2019
![]() User Guide for CORALL RNA-Seq Library Prep Kit with Unique Dual Indices – update 02.12.2020
User Guide for CORALL RNA-Seq Library Prep Kit with Unique Dual Indices – update 02.12.2020
![]() 107MS224V0100 – MSDS Information for UDI 12nt Unique Dual Indexing Add-On Kits – release 17.12.2019
107MS224V0100 – MSDS Information for UDI 12nt Unique Dual Indexing Add-On Kits – release 17.12.2019
![]() 047MS112V0101 – MSDS Information for i5 Dual Indexing Add-On Kits – update 03.07.2018
047MS112V0101 – MSDS Information for i5 Dual Indexing Add-On Kits – update 03.07.2018
![]() Lexogen UDI 12 nt Unique Dual Index Sequences – update 26.03.2021
Lexogen UDI 12 nt Unique Dual Index Sequences – update 26.03.2021
![]() Lexogen i7 and i5 Index Sequences – update 05.05.2020
Lexogen i7 and i5 Index Sequences – update 05.05.2020
![]() i1 12 nt RT-Set for QuantSeq-Pool Sample-Barcode Sequences for Illumina – release 19.08.2020
i1 12 nt RT-Set for QuantSeq-Pool Sample-Barcode Sequences for Illumina – release 19.08.2020
![]() QuantSeq-Pool Calculator – release 05.01.2021
QuantSeq-Pool Calculator – release 05.01.2021
Product Items
| Catalog Nr. | Product Name |
| 107.96 | Lexogen UDI 12 nt Unique Dual Indexing Add-on Kit, Set A1 (UDI12A_0001-0096), 1 rxn/UDI |
| 108.96 | Lexogen UDI 12 nt Unique Dual Indexing Add-on Kit, Set A2 (UDI12A_0097-0192), 1 rxn/UDI |
| 109.96 | Lexogen UDI 12 nt Unique Dual Indexing Add-on Kit, Set A3 (UDI12A_0193-0288), 1 rxn/UDI |
| 110.96 | Lexogen UDI 12 nt Unique Dual Indexing Add-on Kit, Set A4 (UDI12A_0289-0384), 1 rxn/UDI |
| 111.96 | Lexogen UDI 12 nt Unique Dual Indexing Add-on Kit, Set B1 (UDI12B_0001-0096), 1 rxn/UDI |
| 120.384 | Lexogen UDI 12 nt Unique Dual Indexing Add-on Kit, Sets A1-A4 (UDI12A_0001-0384), 1 rxn/UDI |